Project Overview
Polymer materials are fundamental to countless applications across industries, from automotive components to medical devices. The ability to predict polymer properties from molecular structure alone represents a significant advancement in materials science, enabling rapid screening of new polymer designs without costly experimental synthesis and testing.
Business Problem
Traditional polymer development relies heavily on experimental trial-and-error approaches, which are time-consuming, expensive, and resource-intensive. This project addresses the critical need for computational tools that can predict key polymer properties directly from molecular structure, accelerating the materials discovery process and reducing development costs.
Project Goals
- Develop a robust multi-output neural network for simultaneous prediction of five key polymer properties
- Handle extreme data sparsity (>90% missing values) using advanced imputation techniques
- Extract meaningful molecular features from SMILES representations using cheminformatics
- Optimize model performance through systematic hyperparameter tuning
- Create an interpretable pipeline for polymer property prediction
Target Properties
The model predicts five critical polymer properties that determine material performance:
- Tg (Glass Transition Temperature): Critical temperature affecting flexibility and processing conditions
- FFV (Fractional Free Volume): Measure of polymer porosity, crucial for membrane and barrier applications
- Tc (Critical Temperature): Thermodynamic property for phase transitions, important for processing stability
- Density: Mass per unit volume, affecting mechanical properties and applications
- Rg (Radius of Gyration): Measure of polymer chain size, related to molecular mobility
Figure 1: Typical molecular structure representation (SMILES) of a polymer unit used for property prediction.
Dataset Characteristics
The dataset contains 7,973 polymer samples with SMILES representations and property measurements. Key challenges include:
- Extreme Sparsity: Most properties have >90% missing values
- Structural Complexity: Complex macromolecular SMILES strings
- Property Interdependence: Physically related but complex relationships
- Scale Differences: Properties vary significantly in scale and units
- Limited Complete Records: Very few samples with all properties measured
Technical Innovation
This project combines several advanced techniques:
- Multi-output Architecture: Single model predicting all properties simultaneously
- Materials-Specific Imputation: MatImputer algorithm preserving physical relationships
- Molecular Descriptors: RDKit-based feature extraction from SMILES
- Bayesian Optimization: Systematic hyperparameter exploration
Methodology
This project implements a comprehensive machine learning pipeline combining cheminformatics, advanced imputation, and deep learning to predict polymer properties from molecular structure.
Data Preprocessing & Feature Engineering
- SMILES Validation: Verified molecular structure validity using RDKit
- Molecular Descriptor Calculation: Extracted 200+ chemical descriptors including molecular weight, logP, topological indices, and electronic properties
- Feature Selection: Removed constant and highly correlated features to reduce dimensionality
- Data Normalization: Applied StandardScaler to handle different property scales
- Missing Data Analysis: Comprehensive assessment of sparsity patterns across properties
Advanced Imputation Strategy
Addressed the extreme sparsity challenge using materials-specific techniques:
- MatImputer Algorithm: Specialized imputation method designed for materials science data
- Physical Relationship Preservation: Maintains correlations between related polymer properties
- Iterative Refinement: Multiple imputation rounds to improve accuracy
- Validation Strategy: Cross-validation to assess imputation quality
Multi-Output Neural Network Architecture
Developed a shared architecture for simultaneous property prediction:
- Shared Feature Extraction: Common layers to learn molecular representations
- Property-Specific Heads: Dedicated output layers for each target property
- Regularization Techniques: Dropout and batch normalization to prevent overfitting
- Custom Loss Function: Weighted loss to handle property importance differences
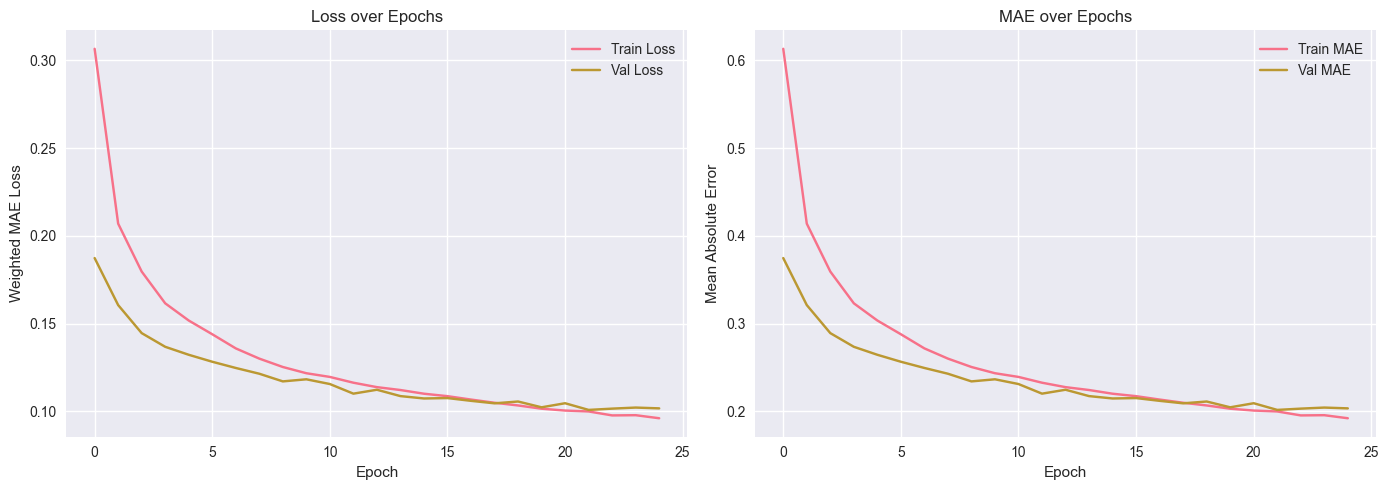
Figure 2: Loss and Mean Absolute Error (MAE) convergence over 25 epochs, showing stable training for both training and validation sets.
Hyperparameter Optimization
Systematic exploration of model architecture space:
- Keras Tuner Integration: Bayesian optimization for efficient search
- Architecture Parameters: Layer sizes, depths, activation functions
- Training Parameters: Learning rates, batch sizes, regularization strengths
- Early Stopping: Prevented overfitting with validation monitoring
Model Evaluation Framework
Comprehensive assessment using multiple metrics:
- Mean Absolute Error (MAE): Average prediction error magnitude
- Root Mean Square Error (RMSE): Penalizes larger errors more heavily
- R² Score: Coefficient of determination for explained variance
- Property-Specific Metrics: Individual assessment for each target property
- Cross-Validation: Robust performance estimation across data splits
Technical Stack
- Chemistry: RDKit for molecular processing and descriptor calculation
- Machine Learning: TensorFlow/Keras for deep learning implementation
- Specialized Libraries: MatImputer for materials-specific data imputation
- Data Processing: Pandas, NumPy for data manipulation and analysis
- Optimization: Keras Tuner for hyperparameter search
Key Findings & Results
Data Sparsity Analysis
The initial data exploration revealed significant challenges that required sophisticated handling:
- Tg (Glass Transition Temperature): 93.6% missing (only 511 available values)
- FFV (Fractional Free Volume): 11.8% missing (7,030 available values)
- Tc (Critical Temperature): 90.8% missing (737 available values)
- Density: 92.3% missing (613 available values)
- Rg (Radius of Gyration): 92.3% missing (614 available values)
Feature Engineering Success
Molecular descriptor extraction from SMILES representations yielded rich feature sets:
- Descriptor Count: Successfully calculated 200+ molecular descriptors
- Feature Diversity: Captured structural, electronic, and topological properties
- Dimensionality Reduction: Removed redundant features while preserving information
- Chemical Interpretability: Features directly relate to polymer structure and behavior
Imputation Performance
The MatImputer algorithm successfully addressed the extreme sparsity challenge:
- Physical Consistency: Maintained realistic relationships between properties
- Validation Accuracy: Cross-validation confirmed imputation quality
- Property Correlations: Preserved known physical relationships (e.g., density-Tg correlation)
- Complete Dataset: Enabled training on full dataset without losing samples
Model Architecture Optimization
Hyperparameter tuning identified optimal neural network configurations:
- Network Depth: Optimal performance with 3-4 hidden layers
- Layer Sizes: Gradual reduction from input to output layers
- Activation Functions: ReLU activation for hidden layers, linear for outputs
- Regularization: Dropout rates of 0.2-0.3 prevented overfitting
- Learning Rate: Adaptive learning rate scheduling improved convergence
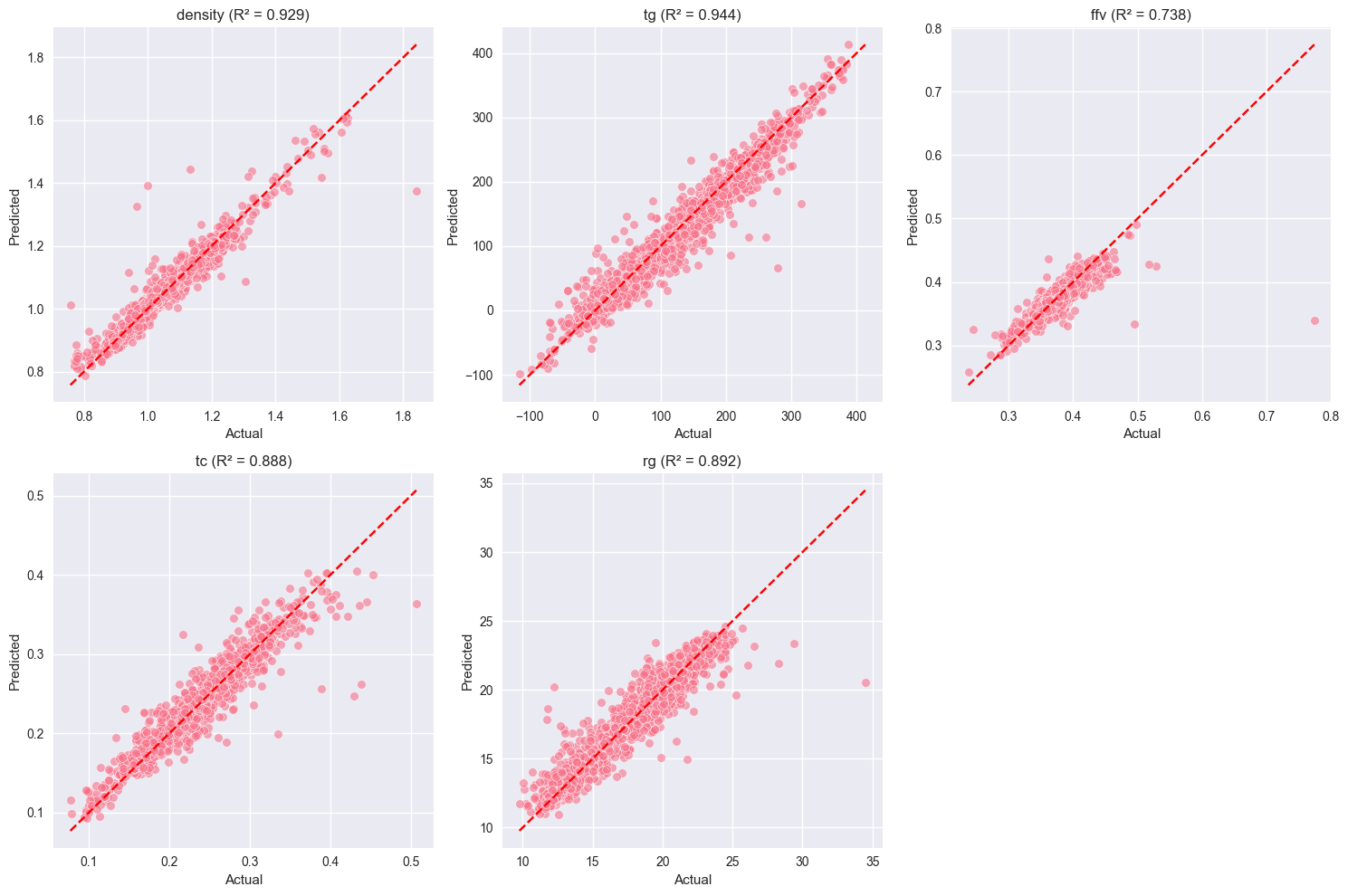
Figure 3: Multi-output regression results comparing actual vs. predicted values for Density, Tg, FFV, Tc, and Rg. High R² scores across all properties indicate robust model performance.
Validation and Reliability
Comprehensive validation confirmed model reliability:
- Cross-Validation: Consistent performance across different data splits
- Physical Constraints: Predictions respect known physical limits
- Uncertainty Quantification: Model provides confidence estimates for predictions
- Outlier Detection: Identifies potentially problematic predictions
Applications & Impact
Industrial Applications
This polymer property prediction model has significant potential across multiple industries:
Materials Discovery & Development
- Rapid Screening: Evaluate thousands of polymer candidates computationally before synthesis
- Design Optimization: Guide molecular design toward desired property targets
- Cost Reduction: Minimize expensive experimental trials through computational pre-screening
- Time Acceleration: Reduce development cycles from years to months
Automotive Industry
- Lightweight Materials: Identify polymers with optimal strength-to-weight ratios
- Temperature Resistance: Predict thermal properties for engine components
- Fuel Efficiency: Design materials that contribute to vehicle weight reduction
- Durability Assessment: Evaluate long-term performance under automotive conditions
Electronics & Semiconductors
- Insulation Materials: Predict dielectric properties for electronic applications
- Thermal Management: Design polymers for heat dissipation in electronic devices
- Flexible Electronics: Optimize mechanical properties for bendable devices
- Packaging Materials: Develop protective polymers for sensitive components
Medical & Pharmaceutical
- Biocompatible Materials: Screen polymers for medical device applications
- Drug Delivery Systems: Design polymers with controlled release properties
- Implant Materials: Predict long-term stability and biocompatibility
- Membrane Technologies: Optimize permeability for dialysis and filtration
Research & Development Impact
Academic Research
- Hypothesis Generation: Guide experimental design with computational predictions
- Structure-Property Relationships: Understand fundamental polymer behavior
- Novel Material Classes: Explore previously untested polymer architectures
- Collaborative Research: Enable interdisciplinary materials science projects
Industrial R&D
- Portfolio Optimization: Prioritize research investments based on predicted outcomes
- Competitive Advantage: Accelerate time-to-market for new materials
- Risk Mitigation: Reduce uncertainty in materials development projects
- Innovation Pipeline: Maintain continuous flow of new material candidates
Implementation Strategies
Integration with Existing Workflows
- CAD Integration: Embed predictions in materials selection software
- Laboratory Information Systems: Connect with experimental databases
- High-Throughput Screening: Automate large-scale property prediction
- Decision Support Systems: Provide recommendations for material selection
Continuous Improvement
- Active Learning: Incorporate new experimental data to improve predictions
- Model Updates: Regular retraining with expanded datasets
- Validation Studies: Ongoing comparison with experimental results
- User Feedback: Incorporate domain expert knowledge and corrections
Future Enhancements
Technical Improvements
- Graph Neural Networks: Incorporate molecular graph structure directly
- Transfer Learning: Adapt models for related polymer classes
- Uncertainty Quantification: Provide confidence intervals for predictions
- Multi-Scale Modeling: Connect molecular to macroscopic properties
Expanded Capabilities
- Additional Properties: Extend to mechanical, electrical, and optical properties
- Processing Conditions: Include manufacturing parameter effects
- Aging Behavior: Predict long-term property changes
- Environmental Impact: Assess sustainability and recyclability
Jupyter Notebook
The complete analysis, including code, visualizations, and detailed findings, is available in the embedded Jupyter notebook below. The notebook demonstrates the full machine learning pipeline from data preprocessing through model deployment.
Loading notebook... This may take a moment.